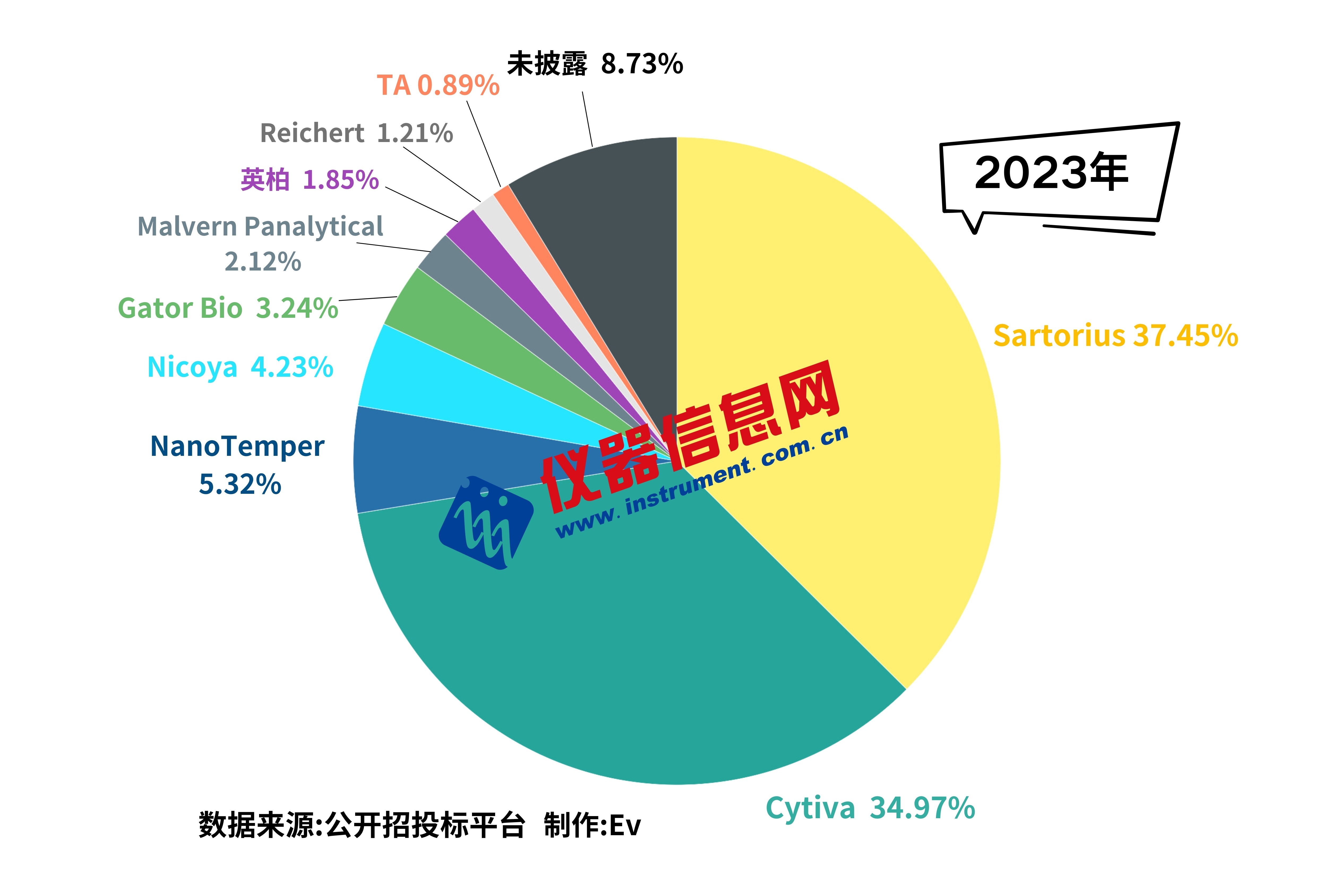
不可思议!诺奖得主David Baker的Binder亲和力有多强?
进入 #重新认识分子互作仪阅读更多话题内容
导读:David Baker课题组已使用Octet®检测蛋白亲和力发表了近80篇文章,其中CNS超过20篇,这里,与大家分享下David Baker教授今年发表的几篇CNS文章。
10月9日,2024年诺贝尔化学奖授予David Baker、Demis Hassabis和John Jumper,以表彰他们在蛋白质设计以及蛋白质结构预测领域做出的杰出贡献。近年来,AI药物迅猛发展,其中,蛋白质从头设计无疑是AI设计药物中的重头戏与热点。该技术不仅能设计针对不可成药靶点的药物和自然中不存在的蛋白质,还大大缩短了药物研发的周期。
被誉为“上帝之手”的华盛顿大学教授David Baker是该领域的佼佼者,发CNS发到手软。当然,AI设计能力再强,也需要通过实验手段去表征设计的蛋白。比如,使用基于生物层干涉技术(BLI)的Octet®非标记分子互作系统检测设计的蛋白与靶点的亲和力。目前,David Baker课题组已使用Octet®检测蛋白亲和力发表了近80篇文章,其中CNS正刊文章就超过20篇!(部分文章列表见文末)
这里,与大家分享下David Baker教授今年发表的几篇CNS文章。
No1. Science-pM级别亲和力的新冠抑制剂[6]
在当时新冠病毒肆虐时,David Baker课题组用计算机软件设计了针对新冠病毒S蛋白的小蛋白抑制剂,通过一系列分子进化以及生化学方法检测后,大部分的抑制剂的中和新冠病毒活性达到了几十pM级别的IC50,其中一个蛋白抑制剂达到11pM的IC50,是当时知道所有新冠抑制剂中最强的!文中进行了两种设计路线:
1) 围绕ACE2与S蛋白结合的α-螺旋结构域设计(方法1)
2) 与S蛋白的RBD结构域对接来设计(方法2)
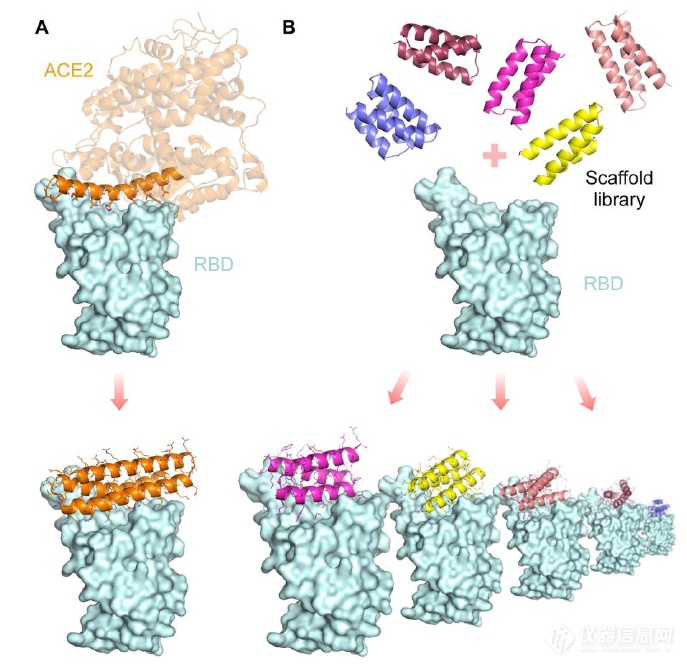
图A为方法1示意图,图B为方法2示意图
技术路线如下:
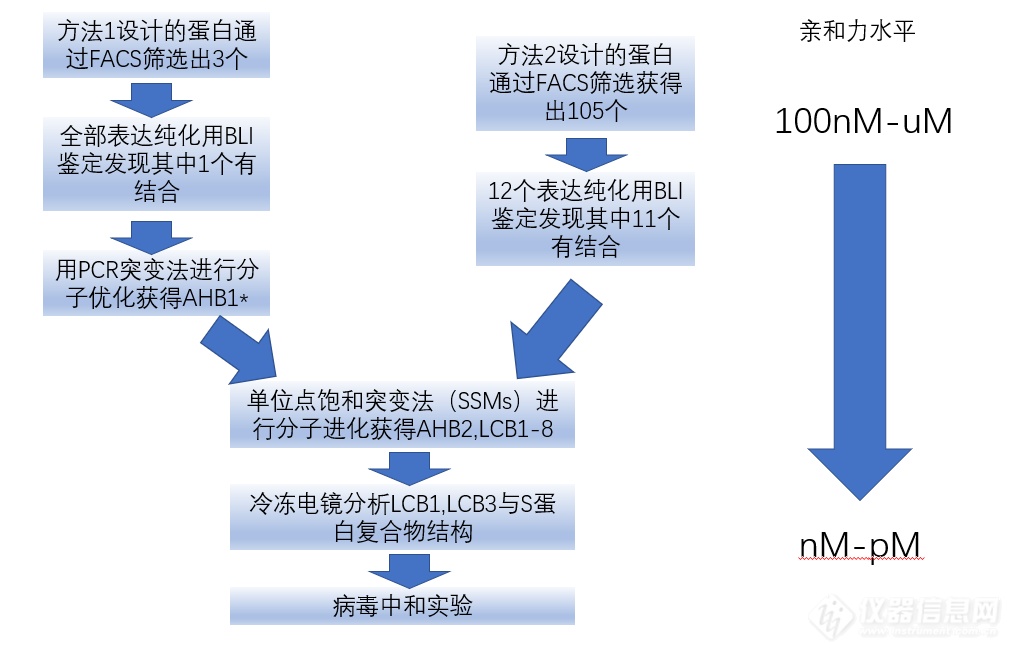
*AHB1亲和力高,但是稳定性差,通过AHB1结构再进行一次设计与筛选,获得AHB2前体。
Octet亲和力测试:
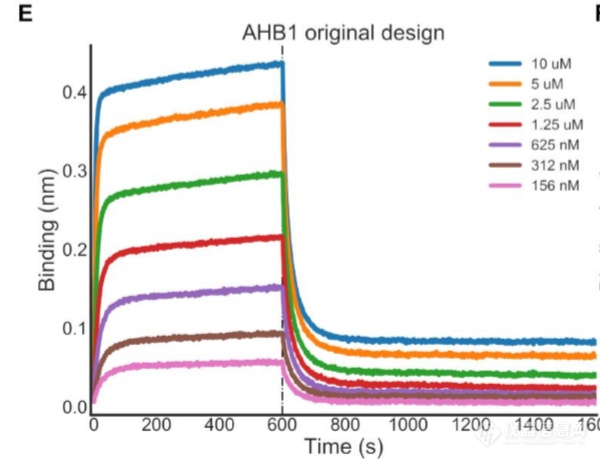
AHB1前体的动力学测定(uM级别亲和力)
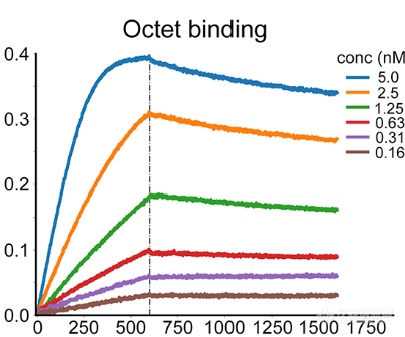
AHB2的动力学测定(nM级别亲和力)
方法2的第一轮筛选的11个候选物(LCD1-8前体)的亲和力为100nM-2uM,而通过分子进化后的多数LCB的亲和力在1-20nM,其中LCB1,LCB3亲和力低于1nM
Octet亲和力测试:
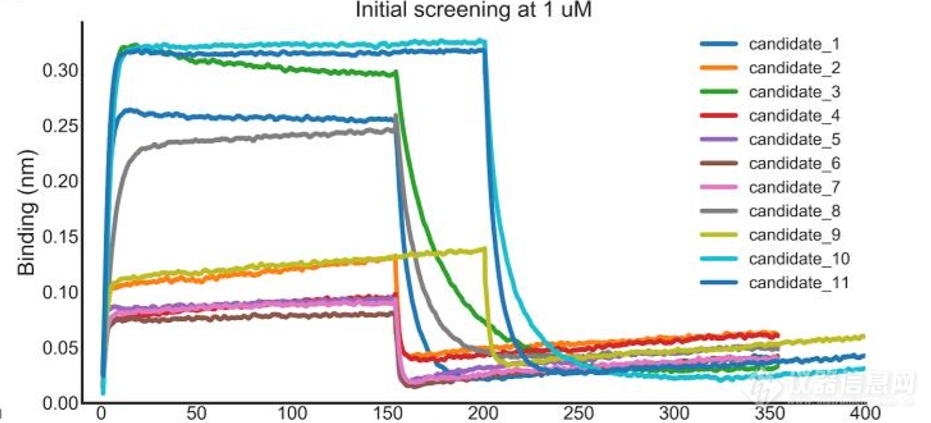
方法2获得的11个候选物的动力学测定,解离都较快,亲和力相对较低;
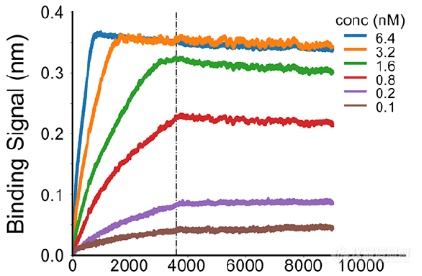
亲和力成熟后的LCB1亲和力测试(低于1nM亲和力)
LCB1也是当时已知中和新冠病毒能力最强的抑制剂。
No.2 Nature—结合扩散模型设计出的高亲和力蛋白[1]
本文结合扩散模型(RF diffusion)和用深度学习算法,实现了从头设计合成蛋白质的目标,设计成功率提高了两个数量级。针对已知的五个靶标,仅需不到100个候选即可达到纳摩尔(nM)级别的亲和力(全部用BLI进行检测)。此外,研究人员还设计了一种蛋白与其底物(流感血凝素)的复合物,并使用冷冻电镜解析其结构。结果显示,冷冻电镜解析的结构与设计的模型几乎完全一致,证明了该模型的准确性。为进一步验证生成的蛋白是否具有结合活性,研究人员继续使用Octet®对这两种蛋白的结合进行验证。
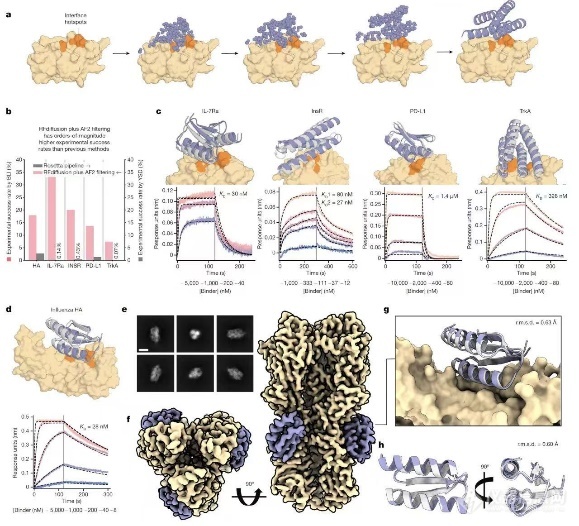
(a-c) 从头设计靶标蛋白的结合蛋白,针对五个靶标,从头设计结合蛋白,BLI 响应信号值≥阳性对照1/2为候选;
(b)RF diffusion成功率高出两个数量级;
(d) 亲和力最高的结合物,结合KD为28nM;
(e-h) 结构学验证
研究团队表示,RF diffusion是对目前蛋白质设计方法的一次综合改进,能够产生总长度达600个氨基酸残基的结构,其复杂性和准确度均超越以往。研究团队还表示,进一步改进该方法或能设计出复杂程度更高的全新蛋白质。
No.3 Nature—超螺旋配对的高亲和力蛋白
针对内在无序区的重复性蛋白和多肽,本文开发了一种基于超螺旋配对的从头设计蛋白的通用性方法。该方法能以大约 20% 的成功率高效设计出多肽结合蛋白,这些蛋白具备低至皮摩(pM)级别的高亲和力、高特异性、高热稳定性以及高精度。此外,该方法还被证实可以靶向更广阔的非重复性多肽区域,并可延伸至人源蛋白的复杂网络。
这种高亲和力、高特异性就是通过Octet®来进行检测的。
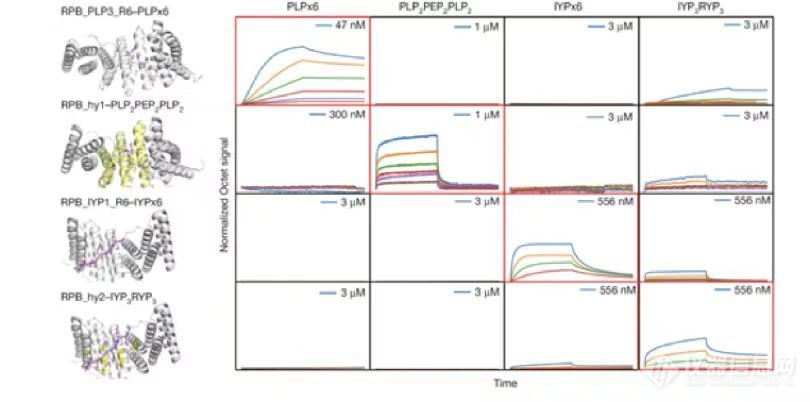
将生物素化的目标肽加载到Octet生物传感器上,并和设计的binder进行结合解离,红色矩形框表示相匹配的结合;可见,设计的binder与目标多肽的良好特异性
三位诺贝尔化学奖得主几乎实现了人类曾经认为不可能的梦想。几十年前,设计全新的蛋白质以及预测蛋白质结构,被视为无法完成的任务。然而,在今天,得益于三位科学家的成果,我们利用人工智能破解了蛋白质结构的密码,这是一场重大的革命。
无论哪个行业都离不开AI,蛋白质设计也一样,尽管蛋白质设计的各种模板和命令还在探索阶段。但无论AI从头设计怎么前沿,相互作用亲和力还是需要通过实验进行验证,蛋白也需要表达纯化才能成为最终产品。所以各位实验室技术大牛们不用怕失业啦!
Octet®为什么如此受欢迎呢?
10月24日,赛多利斯生物分析产品应用科学家杨春兰将为大家分享Octet®非标记分子互作系统技术原理及优势,并对诺奖得主David Baker教授的经典案例进行解读。欢迎参会交流:https://www.instrument.com.cn/webinar/meetings/sartorius241024/ (点击报名)

参考文献:
[1] De novo design of protein structure and function with RFdiffusion
JL Watson, D Juergens, NR Bennett, BL Trippe, J Yim… - Nature, 2023 - nature.com
[2] De novo design of modular peptide-binding proteins by superhelical matching.nature,2023
[3] Top-down design of protein architectures with reinforcement learning.SCIENCEVOL. 380, NO. 6642
[4] Massively parallel de novo protein design for targeted therapeutics
…, X Huang, R Jin, IA Wilson, DH Fuller, D Baker - Nature, 2017 - nature.com
[5] Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing
…, H Kamisetty, P Blair, IA Wilson, D Baker - Nature …, 2012 - nature.com
[6] De novo design of picomolar SARS-CoV-2 miniprotein inhibitors
…, L Stewart, MS Diamond, D Veesler, D Baker - Science, 2020 - science.org
[7] De novo design of bioactive protein switches
…, JE Dueber, WRP Novak, H El-Samad, D Baker - Nature, 2019 - nature.com
[8] High-throughput characterization of protein–protein interactions by reprogramming yeast mating D Younger, S Berger, D Baker
… - Proceedings of the …, 2017 - National Acad Sciences
[9] A potent anti-malarial human monoclonal antibody targets circumsporozoite protein minor repeats and neutralizes sporozoites in the liver
…, R Vistein, C Barillas-Mury, R Amino, D Baker… - Immunity, 2020 - Elsevier
[10] Quadrivalent influenza nanoparticle vaccines induce broad protection
…, MC Crank, L Stewart, KK Lee, M Guttman, D Baker… - Nature, 2021 - nature.com
[11] Transferrin receptor targeting by de novo sheet extension
…, DE Ingber, J Abraham, D Baker - Proceedings of the …, 2021 - National Acad Sciences
[12] Receptor subtype discrimination using extensive shape complementary designed interfaces
…, CJ Kuo, KC Garcia, D Baker - Nature structural & …, 2019 - nature.com
[13] Ultrapotent miniproteins targeting the SARS-CoV-2 receptor-binding domain protect against infection and disease
…, R Ravichandran, L Carter, L Stewart, D Baker… - Cell Host & Microbe, 2021 - Elsevier
[14] Targeting HIV Env immunogens to B cell follicles in nonhuman primates through immune complex or protein nanoparticle formulations
…, G Alter, WR Schief, S Crotty, NP King, D Baker… - npj Vaccines, 2020 - nature.com
[15] De novo design of potent and selective mimics of IL-2 and IL-15
…, GJL Bernardes, M Dougan, KC Garcia, D Baker - Nature, 2019 - nature.com
[16] Computational design of proteins targeting the conserved stem region of influenza hemagglutinin
…, C Dreyfus, JE Corn, EM Strauch, IA Wilson, D Baker - Science, 2011 - science.org
[17] Reconfigurable asymmetric protein assemblies through implicit negative design
…, J Decarreau, HM Morris, A Kang, AK Bera, D Baker - Science, 2022 - science.org
[18] Structural and functional evaluation of de novo-designed, two-component nanoparticle carriers for HIV Env trimer immunogens
…, JP Moore, RW Sanders, NP King, D Baker… - PLoS …, 2020 - journals.plos.org
[19] Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM
…, RF Rocha, ZT Berndsen, D Baker… - Nature …, 2021 - nature.com
[20] Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus
…, KK Lee, D Veesler, CE Correnti, LJ Stewart, D Baker… - Cell, 2019 - Elsevier
[21] Designed protein logic to target cells with precise combinations of surface antigens
…, A Nguyen, S Pun, CE Correnti, SR Riddell, D Baker - Science, 2020 - science.org
[22] De novo design of tyrosine and serine kinase-driven protein switches
…, VH Wysocki, H El-Samad, D Baker - Nature structural & …, 2021 - nature.com
[23] Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site
…, AB Ward, P Yager, DH Fuller, IA Wilson, D Baker - Nature …, 2017 - nature.com
[24] Scaffolding protein functional sites using deep learning
…, F DiMaio, B Correia, S Ovchinnikov, D Baker - Science, 2022 - science.org
[25] Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice
…, BS Freedman, JD Bloom, H Ruohola-Baker… - Science translational …, 2022 - science.org
[26] A computationally designed hemagglutinin stem-binding protein provides in vivo protection from influenza independent of a host immune response
…, IA Wilson, A Dagley, DF Smee, D Baker… - PLoS …, 2016 - journals.plos.org
[27] Computational design of a synthetic PD-1 agonist
…, F DiMaio, KV Tarbell, D Baker - Proceedings of the …, 2021 - National Acad Sciences
[28] First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor
…, SH Orkin, D Baker, H Ruohola-Baker - Proceedings of the …, 2017 - National Acad Sciences
[29] Designed proteins assemble antibodies into modular nanocages
…, H Ruohola-Baker, J Mathieu, D Veesler, D Baker - Science, 2021 - science.org
[30] Designed protein logic to target cells with precise combinations of surface antigens
…, A Nguyen, S Pun, CE Correnti, SR Riddell, D Baker - Science, 2020 - science.org
[31] De novo design of tyrosine and serine kinase-driven protein switches
…, VH Wysocki, H El-Samad, D Baker - Nature structural & …, 2021 - nature.com
[32] Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site
…, AB Ward, P Yager, DH Fuller, IA Wilson, D Baker - Nature …, 2017 - nature.com
[33] Scaffolding protein functional sites using deep learning
…, F DiMaio, B Correia, S Ovchinnikov, D Baker - Science, 2022 - science.org
[34] Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice
…, BS Freedman, JD Bloom, H Ruohola-Baker… - Science translational …, 2022 - science.org
[35] A computationally designed hemagglutinin stem-binding protein provides in vivo protection from influenza independent of a host immune response
…, IA Wilson, A Dagley, DF Smee, D Baker… - PLoS …, 2016 - journals.plos.org
[36] Computational design of a synthetic PD-1 agonist
…, F DiMaio, KV Tarbell, D Baker - Proceedings of the …, 2021 - National Acad Sciences
[37] First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor
…, SH Orkin, D Baker, H Ruohola-Baker - Proceedings of the …, 2017 - National Acad Sciences
[38] Designed proteins assemble antibodies into modular nanocages
…, H Ruohola-Baker, J Mathieu, D Veesler, D Baker - Science, 2021 - science.org
[39] Computational design of a pH-sensitive IgG binding protein
…, SJ Fleishman, D Baker - Proceedings of the …, 2014 - National Acad Sciences
[40] Design of protein-binding proteins from the target structure alone
…, S Bernard, L Stewart, IA Wilson, H Ruohola-Baker… - Nature, 2022 - nature.com
[41] De novo design of modular and tunable protein biosensors
…, J Wi, HJ Hong, L Stewart, BH Oh, D Baker - Nature, 2021 - nature.com
[42] https://www.cell.com /cell/fulltext/s0092-8674(24)00534-8
[43] https://www.nature.com/articles/s41586-024-07948-2
[44] https://www.nature.com/articles/s41589-024-01578-5
[45] https://www.nature.com/articles/s41586-024-07601-y
[46] https://www.nature.com/articles/s41467-024-50919-4
[47] https://www.cell.com/cell/fulltext/S0092-8674(24)00631-7
[48] https://www.nature.com/articles/s41589-024-01718-x
[49] https://www.nature.com/articles/s41422-024-01014-2
来源于:赛多利斯
热门评论
最新资讯
新闻专题