方案详情文
智能文字提取功能测试中
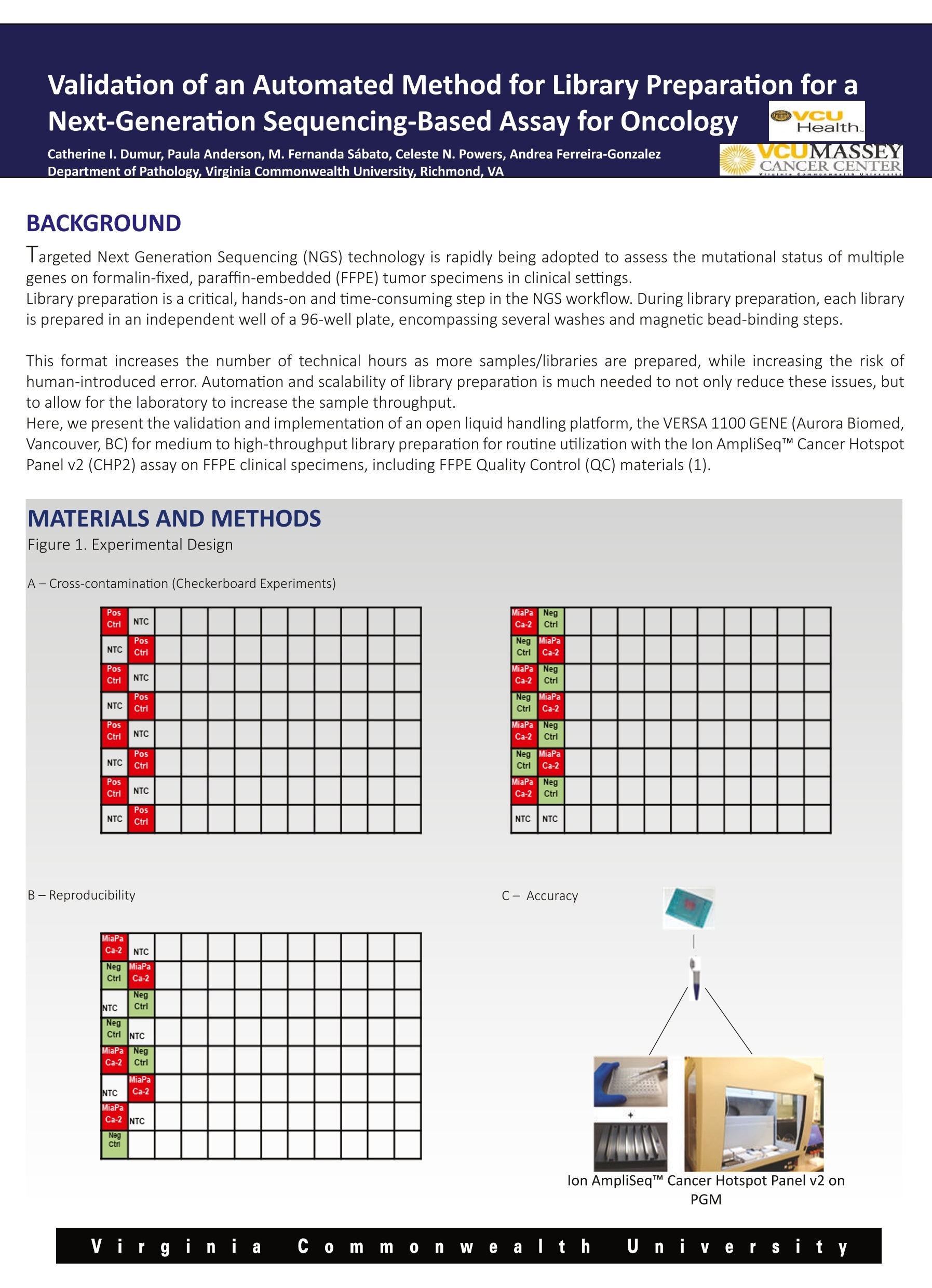
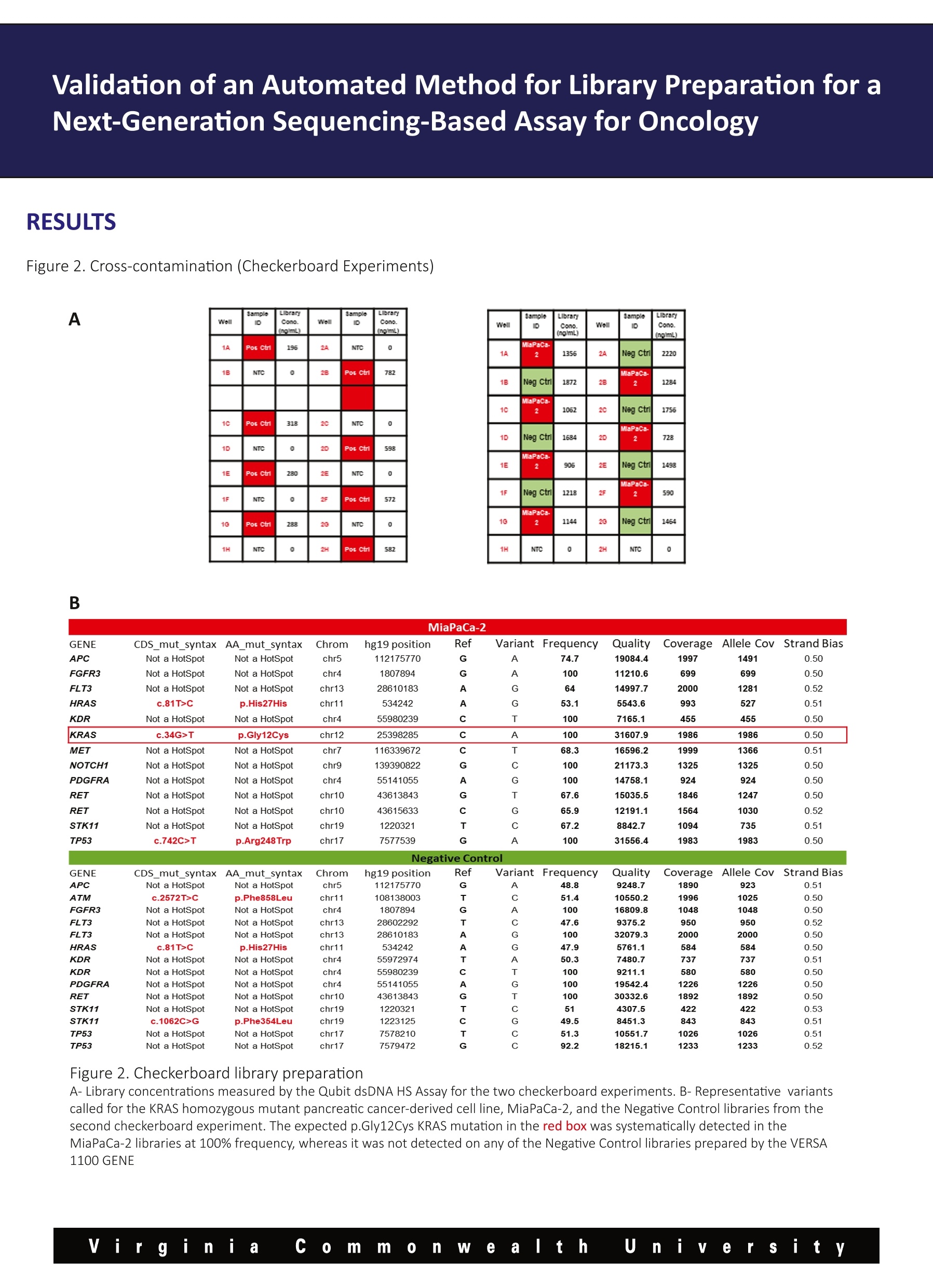
Validation of an Automated Method for Library Preparation for aNext-Generation Sequencing-Based Assay for Oncology 一HVeCalUth. Catherine l. Dumur, Paula Anderson, M. Fernanda Sabato, Celeste N. Powers,Andrea Ferreira-Gonzalez VCUMASSEY Department of Pathology,Virginia Commonwealth University, Richmond, VA CANCER CENTER BACKGROUND laargeted Next Generation Sequencing (NGS) technology is rapidly being adopted to assess the mutational status of multiplegenes on formalin-fixed, paraffin-embedded (FFPE) tumor specimens in clinical settings. Library preparation is a critical, hands-on and time-consuming step in the NGS workflow. During library preparation, each libraryis prepared in an independent well of a 96-well plate, encompassing several washes and magnetic bead-binding steps. This format increases the number of technical hours as more samples/libraries are prepared, while increasing the risk ofnuman-introduced error. Automation and scalability of library preparation is much needed to not only reduce these issues, butto allow for the laboratory to increase the sample throughput. Here, we present the validation and implementation of an open liquid handling platform, the VERSA 1100 GENE (Aurora Biomed,Vancouver, BC) for medium to high-throughput library preparation for routine utilization with the Ion AmpliSeqTM Cancer HotspotPanel v2 (CHP2) assay on FFPE clinical specimens, including FFPE Quality Control (QC) materials (1). Figure 1. Experimental Design A-Cross-contamination (Checkerboard Experiments) Pos Ctrl NTC NTC PosCtrl PosCtrl NTC NTC Pos Ctrl Pos Ctrl NTC NTC PosCtrl PosCtrl NTC NTC Pos ctrl MiaPa Ca-2 NTC NegCtrl MiaPa Ca-2 NTC NegCtrl NegCtrl NTC MiaPa Ca-2 NegCtrl NTC MiaPaCa-2 MiaPa Ca-2 INTC NegCtri PGM Validation of an Automated Method for Library Preparation for aNext-Generation Sequencing-Based Assay for Oncology B MiaPaCa-2 GENE CDS_mut_syntax AA_mut_syntax Chrom hg19 position Ref Variant Frequency Quality/ Coverage Allele Cov Strand Bias APC Not a HotSpot Not a HotSpot chr5 112175770 G A 74.7 19084.4 1997 1491 0.50 FGFR3 Not a HotSpot Not a HotSpot chr4 1807894 G A 100 11210.6 699 699 0.50 FLT3 Not a HotSpot Not a HotSpot chr13 28610183 A G 64 14997.7 2000 1281 0.52 HRAS c.81T>C p.His27His chr11 534242 A G 53.1 5543.6 993 527 0.51 KDR Not a HotSpot Not a HotSpot chr4 55980239 C T 100 7165.1 455 455 0.50 KRAS c.34G>T p.Gly12Cys chr12 25398285 C A 100 31607.9 1986 1986 0.50 MET Not a HotSpot Not a HotSpot chr7 116339672 C T 68.3 16596.2 1999 1366 0.51 NOTCH1 Not a HotSpot Not a HotSpot chr9 139390822 G C 100 21173.3 1325 1325 0.50 PDGFRA Not a HotSpot Not a HotSpot chr4 55141055 A G 100 14758.1 924 924 0.50 RET Not a HotSpot Not a HotSpot chr10 43613843 G T 67.6 15035.5 1846 1247 0.50 RET Not a HotSpot Not a HotSpot chr10 43615633 C G 65.9 12191.1 1564 1030 0.52 STK11 Not a HotSpot Not a HotSpot chr19 1220321 T C 67.2 8842.7 1094 735 0.51 TP53 c.742C>T p.Arg248Trp chr17 7577539 G A 100 31556.4 1983 1983 0.50 Negative Control GENE CDS_mut_syntaxAA mut_syntax Chrom hg19 position Ref Variant Frequency Quality CoverageAllele CovStrand Bias APC Not a HotSpot Not a HotSpot chr5 112175770 G A 48.8 9248.7 1890 923 0.51 ATM c.2572T>C p.Phe858Leu chr11 108138003 T C 51.4 10550.2 1996 1025 0.50 FGFR3 Not a HotSpot Not a HotSpot chr4 1807894 G A 100 16809.8 1048 1048 0.50 FLT3 Not a HotSpot Not a HotSpot chr13 28602292 T C 47.6 9375.2 950 950 0.52 FLT3 Not a HotSpot Not a HotSpot chr13 28610183 A G 100 32079.3 2000 2000 0.50 HRAS c.81T>C p.His27His chr11 534242 A G 47.9 5761.1 584 584 0.50 KDR Not a HotSpot Not a HotSpot chr4 55972974 T A 50.3 7480.7 737 737 0.51 KDR Not a HotSpot Not a HotSpot chr4 55980239 C T 100 9211.1 580 580 0.50 PDGFRA Not a HotSpot Not a HotSpot chr4 55141055 A G 100 19542.4 1226 1226 0.50 RET Not a HotSpot Not a HotSpot chr10 43613843 G T 100 30332.6 1892 1892 0.50 STK11 Not a HotSpot Not a HotSpot chr19 1220321 T C 51 4307.5 422 422 0.53 STK11 c.1062C>G p.Phe354Leu chr19 1223125 C G 49.5 8451.3 843 843 0.51 TP53 Not a HotSpot Not a HotSpot chr17 7578210 T C 51.3 10551.7 1026 1026 0.51 TP53 Not a HotSpot Not a HotSpot chr17 7579472 G C 92.2 18215.1 1233 1233 0.52 Figure 2. Checkerboard library preparation A-Library concentrations measured by the Qubit dsDNA HS Assay for the two checkerboard experiments. B-Representative variantscalled for the KRAS homozygous mutant pancreatic cancer-derived cell line, MiaPaCa-2, and the Negative Control libraries from thesecond checkerboard experiment. The expected p.Gly12Cys KRAS mutation in the red box was systematically detected in theMiaPaCa-2 libraries at 100% frequency, whereas it was not detected on any of the Negative Control libraries prepared by the VERSA 1100 GENE Validation of an Automated Method for Library Preparation for aNext-Generation Sequencing-Based Assay for Oncology RESULTS CONT. Figure 3. Reproducibility A SampleID LibraryprepMethod No. ofVariants Pearson's r (against Manuallibrary prep) SampleID LibraryConc.ng/ml Pos Ctrl Manual 36 N/A NTC 0 Pos Ctrl VERSA1100 36 0.997 PosCtrl 1302 Pos Ctrl VERSA1100 36 0.995 Neg Ctrl 1530 Pos Ctrl VERSA1100 36 0.995 NTC 11.2 Pos Ctrl VERSA1100 36 0.993 Pos Ctrl 746 Pos Ctrl VERSA1100 36 0.994 Neg Ctrl 1564 Neg Ctrl Manual 14 N/A NTC 0 Neg Ctrl VERSA1100 14 0.998 Neg Ctrl VERSA1100 14 0.998 Neg Ctrl VERSA1100 14 0.999 Neg Ctrl VERSA1100 14 0.992 Neg Ctrl VERSA1100 14 Neg Ctrl 0.995 B Figure 3. Reproducibility of Control Samples A-Library concentrations measured by the Qubit dsDNA HSAssay for five Positive and Negative control samples each (Left Panel) andnumber of variants and Pearson’s correlations of variant frequencies with those obtained from manual library preparations (RightPanel). B-Representative curve showing Pearson correlation of the 36 variants frequency identified in the Positive Control sample byboth library preparation methods. Validation of an Automated Method for Library Preparation for aNext-Generation Sequencing-Based Assay for Oncology RESULTS CONT. Figure 4. Accuracy Sample ID No. ofPCRCycles LibraryprepMethod No. ofVariants Pearson's r (againstManual library prep Case 1 20 Manual N/A (libraryfailed) N/A Case 1 23 Manual 19 N/A Case 1 20 VERSA1100 19 0.992 Case_1 23 VERSA1100 19 0.992 Case_2 20 Manual N/A (libraryfailed) N/A Case_2 23 Manual 17 N/A Case_2 20 VERSA1100 17 0.996 Case_2 23 VERSA1100 17 0.997 Case_3 23 Manual 12 N/A Case 3 23 VERSA1100 12 0.995 Figure 4. Accuracy in the variants called on FFPE patient samples Difficult to amplify samples were chosen to compare the library yields and variants called from automatic versus manual librarypreparation protocols were used. Cases 1 and 2 failed to generate libraries using the manual protocol, so they were subjectedto higher number of PCR cycles to generate libraries. For those samples, the VERSA 1100 GENE was used under both conditions,obtaining liberates even at fewer PCR cycles. The number and frequency of the variants found in every case were highlycorrelated. CONCLUSIONS 区 From the checkerboard experiments, we concluded that this automated liquid handling system shows no evidence ofcross-contamination, by either no library on the no template control (NTC) wells, or no variants called on negative samples aftersequencing using the CHP2 assay. 凶 Also, high reproducibility was observed in both, library yields and variants called across all technical replicates of theQuality Control materials. 区 All patient DNA samples yield good quality libraries, including those difficult samples that had previously failed using themanual library preparation method, and variants were called with highly correlated (Pearson’s r>0.990) frequencies to thoseobtained with the manual method. 区 Altogether, our results show that the performance of the VERSATM 1100 Gene automated liquid handling workstation isvery robust and might eliminate human-introduced errors, when compared to the manual library preparation method for theCHP2 assay. Reference 1-Dumur Cl et al. Quality control material for the detection of somatic mutations in fixed clinical specimens by next-generation sequencing. DiagnPathol. 2015;10(1):169. PMID: 26376646, PMCID: PMC4573924 y Vi rginia co m m on e altt h u iiy e r sity Virgini C ommnWealth U n iiversity VERSA™1100基因用于下一代测序分析中靶向文库制备的验证手动文库构建不可扩展,容易出现人为误差。文库制备的自动化是非常必要的,不仅要减少这些问题,而且要允许实验室提高整体样本通量。靶向下一代测序(NGS)技术正迅速应用于评估福尔马林固定石蜡包埋(FFPE)肿瘤标本中多个基因的突变状态。临床环境。在NGS工作流程中,文库准备是一个关键的、实际操作的、耗时的步骤。在库准备过程中,每个库准备在一个96孔板的独立井中,包括几个清洗和磁珠绑定步骤。在2015 American Molecular Pathology meeting会议上,Catherine I.Dumur博士实验室、弗吉尼亚联邦大学分子诊断部副主任兼病理学教授展示了VERSA 1100基因在中高通量文库制备的验证和实现准备常规使用离子AmpliSeq™癌症热点面板v2(CHP2)检测FFPE临床标本,包括FFPE质量控制(QC)材料。Validation of VERSA™ 1100 GENE for Targeted Library Preparation in Next Generation Sequencing-Based AssayManual library preparation is not scalable and is prone to human error. Automation of library preparation is much needed to not only reduce these issues, but to allow for the laboratory to increase overall sample throughput.Targeted Next Generation Sequencing (NGS) technology is rapidly being adopted to assess the mutational status of multiple genes on formalin-fixed, paraffin-embedded (FFPE) tumor specimens inclinical settings. Library preparation is a critical, hands-on and time-consuming step in the NGS workflow. During library preparation, each library is prepared in an independent well of a 96-well plate, encompassing several washes and magnetic bead-binding steps.In this poster, which was presented at the 2015 American Molecular Pathology meeting, the laboratory of Catherine I. Dumur, PhD, Associate Director of Molecular Diagnostics Division and Professor of Pathology at Virginia Commonwealth University, presents the validation and implementation of the VERSA 1100 GENE for medium to high-throughput library preparation for routine utilization with the Ion AmpliSeq™ Cancer Hotspot Panel v2 (CHP2) assay on FFPE clinical specimens, including FFPE Quality Control (QC) materials.information form aurorabiomed.com.cn
关闭-
1/4
-
2/4
还剩2页未读,是否继续阅读?
继续免费阅读全文产品配置单
加拿大欧罗拉生物科技有限公司为您提供《福尔马林固定石蜡包埋(FFPE)肿瘤标本中基因的突变状态检测方案(移液工作站)》,该方案主要用于癌细胞/肿瘤细胞中生化检验检测,参考标准《暂无》,《福尔马林固定石蜡包埋(FFPE)肿瘤标本中基因的突变状态检测方案(移液工作站)》用到的仪器有NGS文库制备 欧罗拉下代测序体系工作站 。
我要纠错
推荐专场
液体处理工作站(移液工作站)
更多相关方案






 咨询
咨询



